Virtual reality-empowered deep-learning analysis of brain cells

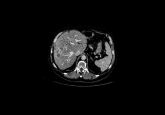
A recent publication in Nature Methods by Ali Ertürk and colleagues (Deep Piction; Munich, Germany) presents DELiVR, an innovative virtual reality-empowered deep-learning pipeline that profoundly enhances the detection and analysis of brain cells in three-dimensional datasets. By investigating cancer-induced changes in brain activity in mice, the study illustrates the practical use of this innovation for three-dimensional medical imaging analysis.
The Technology
At the core of DELiVR is a deep-learning model that operates on data annotated within a virtual reality (VR) environment. The traditional approach to annotating brain images, which typically involves laborious two-dimensional slice-by-slice analysis, is notably time-consuming and prone to errors. DELiVR revolutionizes this process by allowing researchers to annotate volumetric data in a fully immersive three-dimensional space. This not only accelerates the annotation process but also improves the quality of the training data fed into the neural network.
The neural network employed in the present study is a 3D BasicUNet convolutional neural network. Especially well-suited to image data, this model is trained using VR-annotated data. Once trained, it is capable of segmenting brain cells with a high degree of accuracy, outperforming other openly available methods. The output of the neural network is then used by DELVR to reference identified cells against a brain atlas, creating a snapshot of brain-wide activity.
Application- Cancer-induced weight changes in mice
In the current research, DELiVR is demonstrated with a focus on cancer-induced weight changes in mice. It explores how cancer affects brain activity, particularly comparing weight-stable cancer to cachexia, a condition associated with severe weight loss in cancer patients.
The researchers used two different mouse models of colon cancer: one that typically leads to weight-stable conditions (NC26) and another associated with cachexia and weight loss (C26). Both types of cancer cells were transplanted into mice, which were then monitored for changes in body and tumor weight.
Using the DELiVR pipeline, the brains of weight stable mice were found to exhibit increased neural activity in regions associated with sensory processing and foraging behavior, suggesting heightened sensory and motor functions. Conversely, the brains of mice that lost weight were not found to exhibit these increases in activity. It is suggested that this could indicating a possible neurological basis for the lack of compensatory behaviours that could mitigate weight loss.
Implications
The DELiVR pipeline developed for this study contributes a profound improvement in cellular annotation speed and accuracy for researchers across medical imaging domains. Herein it greatly improves the efficiency of this process, which is a notorious bottleneck in research. Additionally, the aforementioned application of DELiVR to cancer illustrates not only the capability of the tool to facilitate detailed brain activity mapping in disease contexts, but also its potential to uncover neurophysiological phenomena. Critically, these insights could subsequently inform scientific and clinical strategies. Finally, the open-source format and high accessibility to researchers creates a strong foundation for application to other medical domains.
Future Directions
The authors suggest that active learning, wherein the deep learning model would react in real time to user annotations and suggests optimal annotation points, represents a clear next step for this innovation. It would facilitate a large jump in efficiency by reducing the number of annotations needed by the user to improve algorithm performance.